Visualisation¶
pyEMAPS provides a set of helper functions to show diffraction patterns. These function are implemented with python’s matplotlib.
si_dif.py and si_bloch.py sample code demonstrate the rendering of diffraction patterns generated for silicon crystal by plotting its kikuchi lines, diffracted beams and HOLZ lines for kinematic simulation and showing the raw image in the case of Bloch simulation.
Users can also access diffraction patterns data directly and visualize the pattern with third party tools such as ImageJ, DigitalMicrograph. as shown in Rendering by Third Party Tools
Builtin Displays¶
Kinematic Diffraction Simulations¶
Additional example of pyEMAPS’s builtin functions for rendering kinematic diffraction simulations for silicon crystal with starting zone changing and camera length set at 2500:

with the following sample code:
from pyemaps import DEF_CBED_DSIZE, DEF_MODE
from pyemaps import EMC
MAX_PROCWORKERS = 4
def generate_difs(name = 'Silicon', mode = DEF_MODE):
from pyemaps import DPList
import concurrent.futures
from pyemaps import Crystal as cryst
cr = cryst.from_builtin(name)
if mode == 2:
dsize = DEF_CBED_DSIZE
zlist=[(0,0,1), (1,1,2), (1,1,1),
(1,3,3), (0,1,2), (0,1,3)]
fs=[]
difs = DPList(name, mode = mode)
emclist =[EMC(zone=z, cl=2500) for z in zlist]
with concurrent.futures.ProcessPoolExecutor(max_workers=MAX_PROCWORKERS) as e:
for ec in emclist:
fs.append(e.submit(cr.generateDP, mode=mode, dsize=dsize, em_controls = ec))
for f in concurrent.futures.as_completed(fs):
try:
emc, diffP = f.result()
difs.add(emc, diffP)
except Exception as e:
print(f'{f} generated an exception: {e}')
exit(1)
difs.sort()
return difs
if __name__ == '__main__':
from pyemaps import showDif
dpl = generate_difs(mode = 2)
showDif(dpl, cShow = True, iShow=True, layout='table', bSave = True)
Dynamic Diffraction Simulations¶
Below is another example of using pyEMAPS builtin display functions to render Bloch simulation images for silicon with:
zone=(1,1,1)
hight voltage vt=100
sample thickness = (500, 1750, 250)
(starting at 500, ending at 1750 with an increment of 250)
sampling points: 40

from pyemaps import EMC, DEF_CBED_DSIZE
MAX_PROCWORKERS = 4
def generate_bloch_images(name = 'Silicon', dsize = DEF_CBED_DSIZE, ckey = 'tilt'):
from pyemaps import Crystal as cryst
from pyemaps import SIMC
cr = cryst.from_builtin(name)
vt = 100
sth = (500, 1750, 250)
simc = SIMC(excitation=(0.3,1.0), bmin=0.1)
try:
bimgs = cr.generateBloch(sampling = 40,
em_controls = EMC(zone=(1,1,1),
vt=vt,
cl=1000,
simc=simc),
sample_thickness = sth
)
except Exception as e:
print(f'Generated an exception: {e}')
return None
bimgs.sort()
return bimgs
if __name__ == '__main__':
from pyemaps import showBloch
imgs = generate_bloch_images()
if imgs is not None:
showBloch(imgs, cShow=True, layout='table', bSave = True)
Stereodiagram¶
Similar build-in function to display stereodiagram output from pyEMAPS is showStereo function in pyEMAPS’s display module. It takes a stereodiagram dictionary objects generated from pyEMAPS’ generateStereo and renders each element by accessing the dictionary objects directly as demonstrated below:
import concurrent.futures
from pyemaps import EMC
MAX_PROCWORKERS = 4
def getStereo(cc, emc = EMC()):
stereo = cc.generateStereo(zone = emc.zone)
return emc, stereo
def generate_stereo_list(name = 'Silicon'):
from pyemaps import Crystal as cr
si = cr.from_builtin(name)
emclist = []
for i in range(-3,3):
emclist.append(EMC(zone=(i,-i,1)))
fs = []
slist=[]
with concurrent.futures.ProcessPoolExecutor(max_workers=MAX_PROCWORKERS) as e:
for ec in emclist:
fs.append(e.submit(getStereo, si, emc=ec))
for f in concurrent.futures.as_completed(fs):
try:
emc, stereo = f.result()
except Exception as e:
print('failed to generate stereodiagram with ' + str(e))
exit(1)
else:
slist.append((emc, stereo))
slist.sort(key=lambda x: x[0])
return slist
if __name__ == '__main__':
from pyemaps import showStereo
stereoList = generate_stereo_list()
showStereo(stereoList,
name='Silicon',
cShow = True,
iShow=True,
zLimit = 1,
layout='table')

Pyemaps GUI Web Service¶
Driven by the latest pyEMAPS, a web server is setup for those who are interested in better visualization of pyEMAPS simulations and calculations. This service is for a nominal subscription fees. Go to https://emaps.emlabsolutions.com to sign up.
Below is an example of the ineractive 3-d display of atomic structure through Jmole based on pyEMAPS crystal structure calculations:

Rendering by Third Party Tools¶
pyEMAPS kinematic diffraction simulation results are captured by DP or diffPattern class:
from pyemaps import Crystal as cr
si = cr.from_builtin('Silicon')
_, dp = si.generateDP()
dp.klines # Kikuchi lines list
dp.nklines # number of Kikuchi lines, same as len(dp.klines)
dp.disks # diffracted beams list
dp.ndisks # number of diffracted beams, same as len(dp.disks)
dp.hlines # HOLZ lines list
dp.nklines # number of HOLZ lines, same as len(dp.hlines)
where dp.klines is a python list of Line class objects and dp.disks of Disk class objects and so on. Each of the objects is accessible for its own data such as points when plotting.
Below is an exmaple of rendering a diffraction pattern generated for Diamond builtin crystal in DigitalMicrograph:
Note
For complete python script integrating pyEMAPS into DM, see si_stereo.py in samples directory.
def show_diffract(dp, md=1, name = 'Diamond'):
from pyemaps import XMAX, YMAX
import DigitalMicrograph as DM
shape = (2*XMAX*mult,2*YMAX*mult)
#A new image from numpy array and initilize it to black background
dif_raw = np.ones((shape), dtype = np.float32)
dif_raw[:,:] = 255.0
#DM create the image
dm_dif_img = DM.CreateImage(dif_raw)
dif_img = dm_dif_img.ShowImage()
dif_img_disp = dm_dif_img.GetImageDisplay(0)
#validate diffraction mode
if md <1 or md > 2:
print(f'diffraction mode provided {md} not supported')
return 1
#set image title
img_title = str(f'Kinematic Diffraction Simulation: {name} in {DIFF_MODE[md-1]} Mode')
dm_dif_img.SetName(img_title)
#plotting Kikuchi and HOLZ lines and spots as DM components
num_klines = dp.nklines
if num_klines > 0:
klines = dp.klines
for kl in klines:
x1,y1,x2,y2, inten = kl #inten: intensity
xx1, yy1, = (x1+ XMAX)*mult,(y1 + YMAX)*mult
xx2, yy2 = (x2 + XMAX)*mult,(y2 + YMAX)*mult
kline = dif_img_disp.AddNewComponent(2, xx1, yy1, xx2, yy2)
SetCommonProp(kline)
if inten/5 > 0.8:
kline.SetForegroundColor(0.3, 0.3, 0.3) # dark grey
elif inten/5 > 0.6:
kline.SetForegroundColor(0.6, 0.6, 0.6)
elif inten/5 > 0.4:
kline.SetForegroundColor(0.8, 0.8, 0.8)
else:
kline.SetForegroundColor(0.9, 0.9, 0.9) # light grey
kline.SetBackgroundColor(0.2,0.2,0.5)# dark blue
num_disks = dp.ndisks
if num_disks > 0:
disks = dp.disks
for d in disks:
x1, y1, r, i1, i2, i3 = d
xx, yy, rr = (x1 + XMAX)*mult, (y1 + YMAX)*mult, r*mult
idx = '{:d} {:d} {:d}'.format(i1,i2,i3)
disk = dif_img_disp.AddNewComponent(6, xx-rr, yy-rr, xx+rr, yy+rr)
SetCommonProp(disk)
disk.SetForegroundColor(0.0,0.0,1.0) # blue
disk.SetBackgroundColor(0.5,0.5,0.75)# dark blue
if md == 1:
disk.SetFillMode(1)
else:
disk.SetFillMode(2)
# a bit tricky to figure out the index location, has to use
# the proxy component indxannot0 first.
indxannot0 = DM.NewTextAnnotation(0, 0, idx, 10)
t, l, b, r = indxannot0.GetRect()
w = r-l
h = b-t
nl = xx - ( w / 2)
# nr = xx + ( w / 2)
nt = yy -rr - h if md ==1 else yy - (h / 2)
# nb = yy + rr + h if md == 1 else yy + (h / 2)
indxannot = DM.NewTextAnnotation(nl, nt, idx, 10)
dif_img_disp.AddChildAtEnd(indxannot)
SetCommonProp(indxannot)
indxannot.SetForegroundColor(0.9,0,0) #light red
indxannot.SetBackgroundColor(1,1,0.5)
if md == 2:
num_hlines = dp.nhlines
if num_hlines > 0 :
hlines = dp.hlines
for hl in hlines:
x1, y1, x2, y2, inten = hl
xx1, yy1 = (x1 + XMAX)*mult, (y1 + YMAX)*mult
xx2, yy2 = (x2 + XMAX)*mult, (y2 + YMAX)*mult
hline = dif_img_disp.AddNewComponent(2, xx1, yy1, xx2, yy2)
SetCommonProp(hline)
if inten/5 > 0.8:
hline.SetForegroundColor(0.1,0.7,0.3) # dark grey
elif inten/5 > 0.6:
hline.SetForegroundColor(0.1,0.7,0.6)
elif inten/5 > 0.4:
hline.SetForegroundColor(0.1,0.7,0.8)
else:
hline.SetForegroundColor(0.1,0.7,0.9) # light grey
del dm_dif_img
return 0
where dp is the kinematic diffraction pattern generated from generateDP call.
Note
To get the above display work in DM, pyEMAPS must be installed in DM’s python environment.
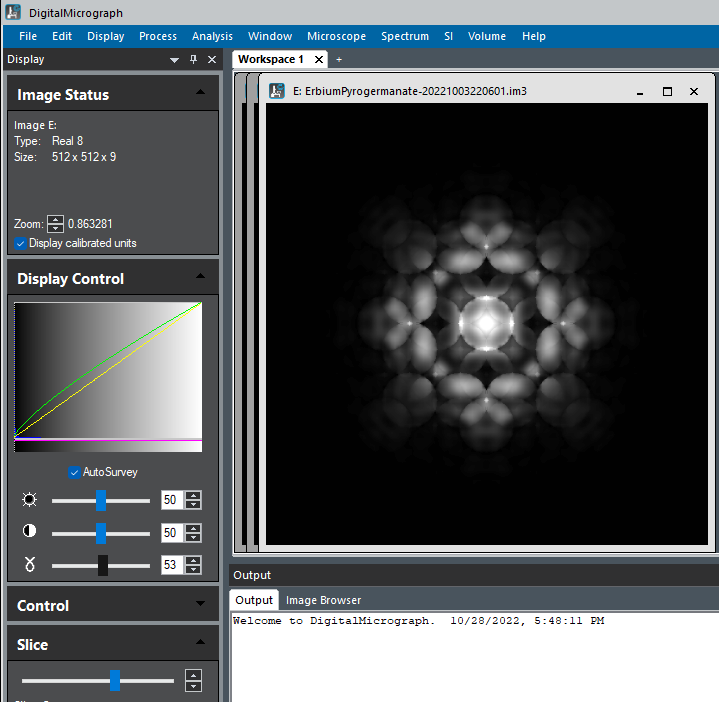
Dynamic Diffraction Patterns¶
The output from bloch dynamic diffraction simulation is an array of 2 or 3 dimensional floating numbers representing image intensities, depending on whether one or multiple slices of dynamic diffraction images are generated.
pyEMAPS provides an option for users to save the images as raw image files that can be imported into ImageJ or DigitalMicrograph. Refer to generateBloch or getBlochImages for details.
Below is a visualization of a raw bloch images files saved from a pyEMAPS dynamic simulation run in Digita;Micrograph. The raw image contains 10 images each at a sample thickness range betwwen 0 and 1000.
Constructing Crystal¶
pyEMAPS’ generateMxtal function produces .XYZ file that can be improted into Jmole and visualize the crystal in 3D atomic structure. The following is an example of such rendering:
